Detection of Active Binding Sites with the Combination of DNase Hypersensitivity and Histone Modifications
Eduardo Gade Gusmão, Christoph Dieterich, Martin Zenke and Ivan Gesteira Costa.
Method
We propose an HMM-based approach to integrate both DNase I hypersensitivity and histone modifications for the detection of open chromatin regions and active binding sites.
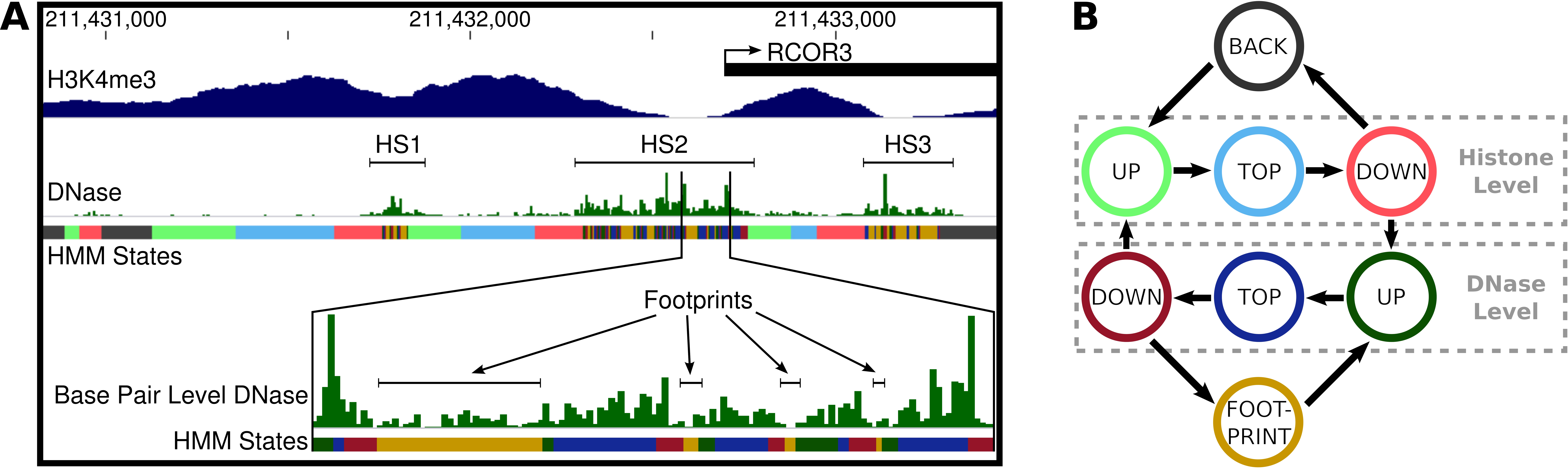
Within transcription factor binding sites (TFBSs), there is a specific grammar of DNase I digestion and histone marks (see A in figure above). We have therefore devised a multivariate HMM (see B in figure above) to model this regulatory grammar by simultaneous analysis of DNase-seq and the ChIP-seq profiles of H3K4me3 (indicative of promoters) or H3K4me1 (indicative of enhancers) on a genome-wide level.
The HMM has as input a normalized and a slope signal of DNase-seq and one of the histone marks. It can therefore detect the increase, top and decrease regions of either histone modification and DNase signals. The genomic regions annotated with the ‘footprint’ HMM state are considered our predictions and represent likely binding sites within that cell’s context.
The full description can be found in the publication.
Software
The tool HINT (Hmm-based IdeNtification of Tf footprints) is available as part of the Regulatory Genomics Toolbox. Inside the tool you can find the option to create footprints based on DNase-seq and ChIP-seq for histone modifications.
Additionally, in the website above you can find sample scripts and the complete manual with detailed instructions allowing some experimental flexibility.
Datasets
We make available experimental benchmarking data in the following link.
The compressed folder contains data concerning:
- ChIP-seq peaks for transcription factors tested (in order to create the evaluation datasets).
- Footprints for all methods: Boyle, Cuellar, Centipede, DH-HMM, H-HMM and Neph.
- HMM models.
- Transcription factors binding sites obtained with motif matching (in order to create the evaluation datasets).
Citing
If you use HINT in your research, we kindly ask you to cite the following publication:
Gusmao EG, Dieterich C, Zenke M and Costa IG. “Detection of Active Transcription Factor Binding Sites with the Combination of DNase Hypersensitivity and Histone Modifications”. Bioinformatics, 30(22):3143-3151, 2014. [Full Text]
Bibtex:
@article{gusmao2014,
author = {Gusmao, Eduardo G. and Dieterich, Christoph and Zenke, Martin and Costa, Ivan G.},
citeulike-article-id = {13340527},
day = {15},
doi = {10.1093/bioinformatics/btu519},
issn = {1460-2059},
journal = {Bioinformatics},
keywords = {dnase1, histone\_marks, tfbs},
month = nov,
number = {22},
pages = {3143--3151},
pmid = {25086003},
posted-at = {2014-08-29 05:42:07},
priority = {2},
publisher = {Oxford University Press},
title = {Detection of active transcription factor binding sites with the combination of {DNase} hypersensitivity and histone modifications},
url = {http://dx.doi.org/10.1093/bioinformatics/btu519},
volume = {30},
year = {2014}
}